ASTM D5465-93(1998)
(Practice)Standard Practice for Determining Microbial Colony Counts from Waters Analyzed by Plating Methods
Standard Practice for Determining Microbial Colony Counts from Waters Analyzed by Plating Methods
SCOPE
1.1 These practices cover recommended procedures for counting colonies and reporting colony-forming units (CFU) on membrane filters (MF) and standard pour and spread plates.
1.2 This standard does not purport to address all of the safety problems, if any, associated with its use. It is the responsibility of the user of this standard to establish appropriate safety and health practices and determine the applicability of regulatory limitations prior to use.
General Information
Relations
Standards Content (Sample)
NOTICE: This standard has either been superseded and replaced by a new version or withdrawn.
Contact ASTM International (www.astm.org) for the latest information
An American National Standard
Designation: D 5465 – 93 (Reapproved 1998)
Standard Practice for
Determining Microbial Colony Counts from Waters Analyzed
by Plating Methods
This standard is issued under the fixed designation D 5465; the number immediately following the designation indicates the year of
original adoption or, in the case of revision, the year of last revision. A number in parentheses indicates the year of last reapproval. A
superscript epsilon (e) indicates an editorial change since the last revision or reapproval.
1. Scope
1.1 These practices cover recommended procedures for
counting colonies and reporting colony-forming units (CFU)
on membrane filters (MF) and standard pour and spread plates.
1.2 This standard does not purport to address all of the
safety problems, if any, associated with its use. It is the
responsibility of the user of this standard to establish appro-
priate safety and health practices and determine the applica-
bility of regulatory limitations prior to use.
2. Significance and Use
2.1 These practices provide a uniform set of counting,
calculating, and reporting procedures for ASTM test methods
in microbiology.
Section
A—Counting Colonies on Membrane Filters 4
B—Counting Colonies on Pour Plates 5
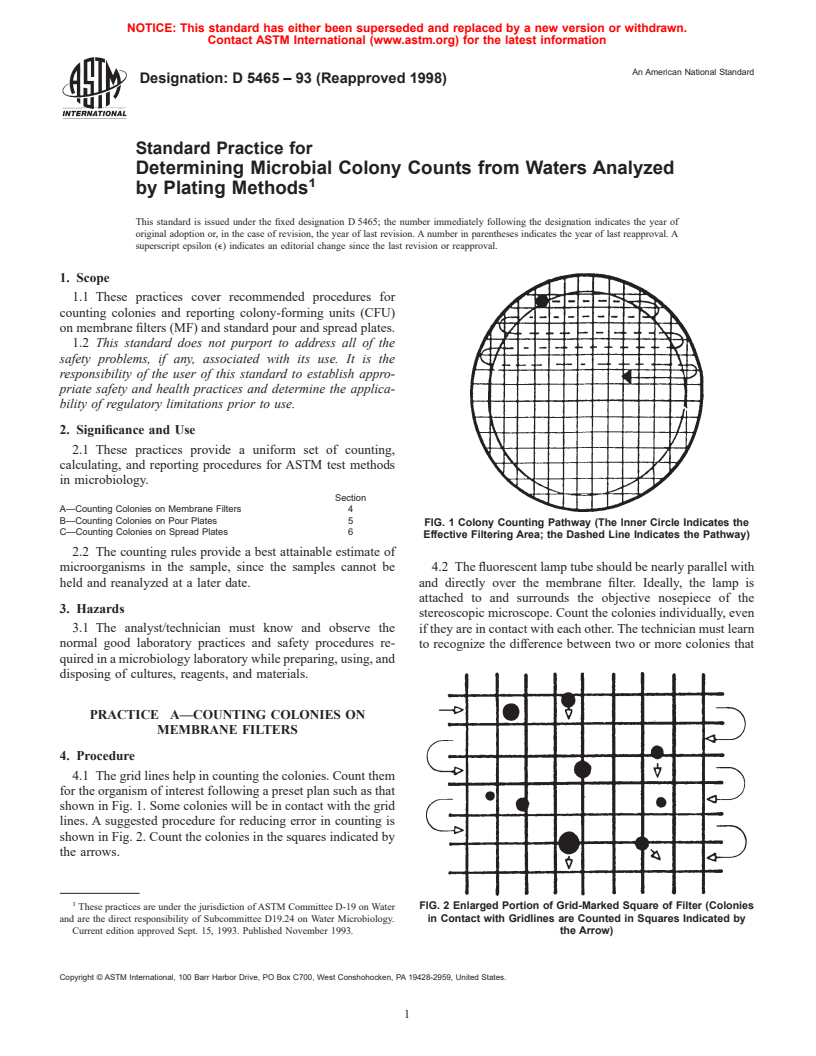
FIG. 1 Colony Counting Pathway (The Inner Circle Indicates the
C—Counting Colonies on Spread Plates 6
Effective Filtering Area; the Dashed Line Indicates the Pathway)
2.2 The counting rules provide a best attainable estimate of
microorganisms in the sample, since the samples cannot be 4.2 The fluorescent lamp tube should be nearly parallel with
held and reanalyzed at a later date. and directly over the membrane filter. Ideally, the lamp is
attached to and surrounds the objective nosepiece of the
3. Hazards
stereoscopic microscope. Count the colonies individually, even
3.1 The analyst/technician must know and observe the
if they are in contact with each other. The technician must learn
normal good laboratory practices and safety procedures re- to recognize the difference between two or more colonies that
quired in a microbiology laboratory while preparing, using, and
disposing of cultures, reagents, and materials.
PRACTICE A—COUNTING COLONIES ON
MEMBRANE FILTERS
4. Procedure
4.1 The grid lines help in counting the colonies. Count them
for the organism of interest following a preset plan such as that
shown in Fig. 1. Some colonies will be in contact with the grid
lines. A suggested procedure for reducing error in counting is
shown in Fig. 2. Count the colonies in the squares indicated by
the arrows.
FIG. 2 Enlarged Portion of Grid-Marked Square of Filter (Colonies
These practices are under the jurisdiction of ASTM Committee D-19 on Water
and are the direct responsibility of Subcommittee D19.24 on Water Microbiology. in Contact with Gridlines are Counted in Squares Indicated by
Current edition approved Sept. 15, 1993. Published November 1993. the Arrow)
Copyright © ASTM International, 100 Barr Harbor Drive, PO Box C700, West Conshohocken, PA 19428-2959, United States.
D 5465 – 93 (1998)
have grown into contact with each other and the single, dently to final reporting units, and then average for the final
irregularly shaped colonies that sometimes develop on mem- reported value. For example, assume that volumes of 0.3, 0.1,
brane filters. The latter colonies are usually associated with a 0.03, and 0.01 mL produced colony counts of too numerous to
fiber or particulate material and conform to the shape and size count (TNTC), 75, 30, and 8, respectively. In this example, two
of the fiber or particulates. Colonies that have grown together volumes, 0.1 and 0.03, produce colonies in an acceptable
almost invariably show a very fine line of contact. counting range. Carry each MF count independently to a
4.3 Count the colonies with a stereoscopic (dissecting) count/mL or count/100 mL:
microscope that provides a magnification of at least 10 to 153.
3 1 ~or 3 100! 5 750 CFU/mL ~or 75 000 CFU/100 mL!
4.4 See Table 1 for guidance on acceptable counting limits.
0.1
4.5 Calculation of Results—Select the membrane with the (6)
number of CFU in the acceptable range and calculate the
3 1 ~ or 3 100!
count/reporting volume according to the following general
0.03
formulae:
= 1 000 CFU/mL (or100 000 CFU/100 mL)
colonies counted
Then calculate the arithmetic mean of these counts to obtain
CFU/mL 5 3 1 (1)
volume of sample filtered in mL
the final reported value:
750 1 1 000
colonies counted 5 875 (7)
CFU/100 mL 5 3 100 (2)
volume of sample filtered in mL
Report as 880 CFU/mL.
4.6 Counts Within the Acceptable Limits:
75 000 1 100 000
4.6.1 The acceptable range of counts on a membrane for
5 87 500 (8)
samples that are diluted is a function of the organism/test
combination as given in Table 1. Report as 88 000 CFU/100 mL.
4.6.5 For finished drinking water samples only, countable
4.6.2 Assume that the filtration of volumes of 80, 20, 5, and
1 mL produced counts of 250, 60, 15, and 4, respectively. Do membranes may contain from one colony to the upper limit of
the test (see Table 1). Count the target colonies/volume filtered.
not count the colonies on all filters. Select the MF(s) within the
acceptable counting range and then limit the actual counting to Calculate and report the number of CFU/100 mL.
4.7 Counts Outside Acceptable Limits:
such membranes. After selecting the best MF for counting, in
this case that with a 60-CFU count, the analyst counts CFU 4.7.1 Zero counts recorded as < values/volume filtered are
acceptable for sample volumes of 100 mL or more.
according to the procedures shown in Fig. 1 and Fig. 2 and
applies the general formula as follows: 4.7.2 If full-volume samples are filtered, such as 25, 50, or
100 mL, and the resulting count is 1 to 19 colonies, these
3 1 ~or 3 100! 5 3 ~or 300! (3)
values are acceptable although <20. The count is adjusted to 1
or 100 mL for reporting. For example, a count of 1 colony/25
Report as 3 CFU/mL or 300 CFU/100 mL.
mL is adjusted:
4.6.3 If there are acceptable counts on replicate plates, carry
the counts independently to final reporting units, and then
3 1 ~or 3 100!5,1 ~or 4! (9)
calculate the arithmetic mean of these counts to obtain the final
reported value. For example, 1 mL volumes produced counts of
Report as <1 CFU/mL (or 4 CFU/100 mL).
26 and 36 CFU/mL or counts of 2600 and 3600 CFU/100 mL: 4.7.3 If all MF counts are <20/volume filtered, select the
most nearly acceptable count (for non-drinking waters). For
26 1 36
5 31 (4)
example, assume a count in which sample volumes of 1, 0.3,
and 0.01 mL produced CFU counts of 14, 3, and 0, respec-
Report as 31 CFU/mL.
tively. No CFU count falls within recommended limits here.
2600 1 3600
Calculate on the basis of the most nearly acceptable plate
5 3100 (5)
count, 14, and report with a qualifying remark:
Report as 3100 CFU/100 mL.
3 1 ~ or 3 100! 5 14 ~ or 1400! (10)
4.6.4 If more than one dilution produced acceptable counts,
1.0
count the colonies for each dilution, carry the counts indepen-
Report as estimated count: 14 CFU/mL (or 1400 CFU/100
mL).
TABLE 1 Recommended Counting Range for High-Density
4.7.4 If counts from all membranes are zero, calculate using
A
Samples
the count from largest filtration volume. For example, sample
Microorganism Colony Count Remarks
volumes of 25, 10, and 2 mL produced CFU counts of 0, 0, and
Total coliform bacteria, MF, 47 mm 20 to 80 Upper limit, 200
0, respectively, and no actual calculation is possible, even as an
colonies of all types
estimated report. Calculate the number of CFU per 100 mL that
Fecal coliform bacteria, MF, 47 mm 20 to 60
Fecal streptococci, MF, 47 mm 20 to 100
would have been reported if there had been one CFU on the
Heterotrophic spread plate count 20 to 200
filter representing the largest filtration volume; thus:
Heterotrophic pour plate count 30 to 300 Upper limit, 300
A ,1
Colo
...







Questions, Comments and Discussion
Ask us and Technical Secretary will try to provide an answer. You can facilitate discussion about the standard in here.